Biological Science Faculty Member
Dr. Qian Yin
- Office: 506 Kasha Laboratory
- Office: 850-644-1747
- Area: Cell and Molecular Biology
- Lab: Kasha Laboratory
- Lab: 850-644-2286
- E-mail: yin@bio.fsu.edu

Assistant Professor
Ph.D., Weill Cornell Medical College, 2008
Graduate Faculty Status
Research and Professional Interests:
My lab is interested in how individual proteins, or protein assemblies, mediate biological processes in the linked areas of innate immunity, inflammation and host-pathogen interactions. Longer-term perspectives extend to the interplay between inflammation, infection, autophagy, and cytoskeleton rearrangement. We study the mechanisms of pathogen-associated molecular patterns (PAMPs) recognition, viral restriction, evasion or suppression of host immunity and their intricate network using a combination of biochemical, structural (including X-ray crystallography and cryo-EM), cellular and pharmacological approaches.
Mechanistic and structural studies of IFN-inducible GTPases
GTPases have been meticulously documented for their roles to regulate multiple cellular processes spanning from cell mobility, membrane fusion and fission, to cytokinesis and vesicle transport. IFN-inducible GTPases, consisting of more than forty members in human and mice, are conserved from vertebrates to human. They are among the most highly expressed ISGs, sometimes accounting for twenty percent of all proteins induced by IFN-γ. Yet the functions of IFN-inducible GTPases remain largely unknown.
IFN-inducible GTPases can be classified into three major subfamilies: guanylate-binding proteins (GBPs), immunity-related GTPases (IRGs), and myxovirus resistance (Mx) proteins. IRGs can be further divided into GKS and GMS IRGs based on sequence features. They are all part of cell-autonomous resistance against microbial infections by patrolling the intracellular space, targeting pathogen-containing vacuoles (PVs) and viruses, promoting inflammasome formation, and coordinating assembly of oxidative and autophagic complexes for wide-spectrum pathogens elimination. 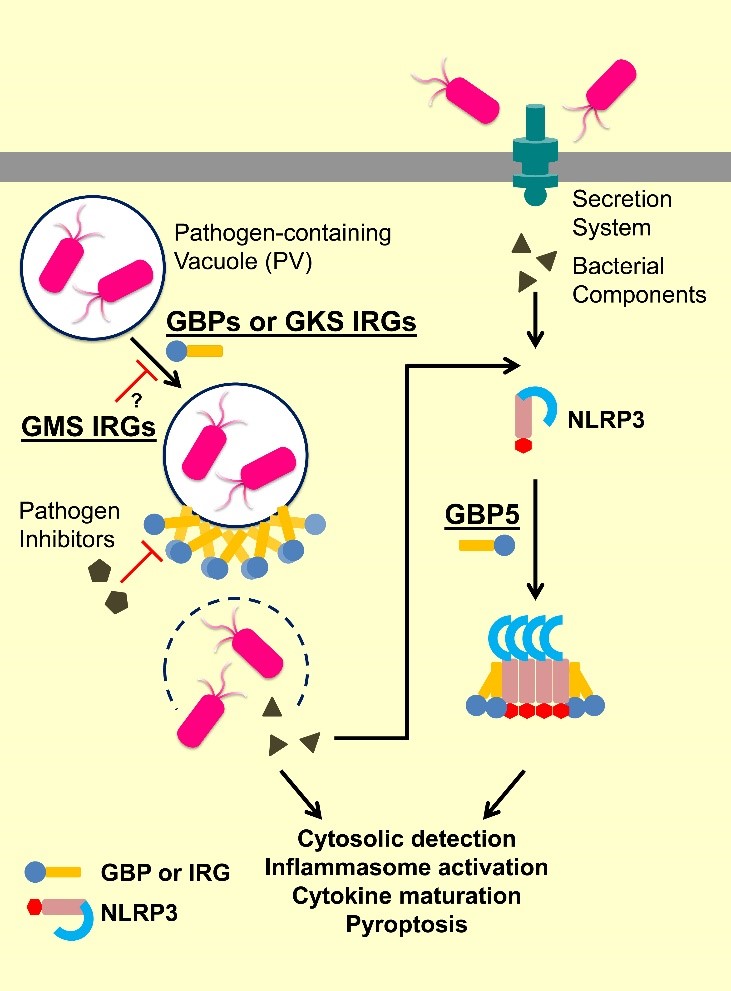
I propose to study the working mechanisms and structure-function relationships of GBPs and IRGs.
1. Elucidating GBP self-organization by biochemical, structural and microscopic methods
2. Understanding the specificity of GBPs and IRGs towards foreign entities
3. Revealing the pathogen counterattack strategies
Highly motivated individuals are welcome to contact Dr. Yin for positions at all levels: undergraduate student researchers, graduate students, postdoctoral fellows, and technicians.
Selected Publications:
For a complete list of publication, please see
https://www.ncbi.nlm.nih.gov/myncbi/10SndZqACg8/bibliography/public/
Bhattacharya M, Bhowmik D, Tian Y, He H, Zhu F, and Yin Q. The Dengue virus protease NS2B3 cleaves cyclic GMP-AMP synthase to suppress cGAS activation. J Biol Chem. 2023; 299(3):102986 [DOI] [PMC10011430]
Tian Y, Datta I, Yang R, Wan C, Wang B, Crisman L, He H, Brautigam CA, Li S, Shen J*, and Yin Q*. Oligomer-to-monomer transition underlies the chaperone function of AAGAB in AP1/AP2 assembly. Proc Natl Acad Sci USA. 2023; 120(2):e2205199120 [PMC9926252]
Bhowmik D, Tian Y, Wang B, Zhu F*, and Yin Q*. Structural basis of higher order oligomerization of KSHV inhibitor of cGAS. Proc Natl Acad Sci USA. 2022; 119(33): e2200285119 [PMC35939686]
Bhowmik D, Du M, Tian Y, Ma S, Wu J, Chen Z, Yin Q, and Zhu F. Cooperative DNA binding mediated by KicGAS/ORF52 oligomerization allows inhibition of DNA-induced phase separation and inhibition of cGAS. Nucleic Acids Res. 2021; 49(16): 9389-9403 [PMC8450086]
Tian Y, Yin Q, Structural analysis of the HIN1 domain of interferon-inducible protein 204. Acta Crystallogr F Struct Biol Commun. 2019; 75 (Pt6): 455-60 [PMC6572094]
Wang B, Yin Q, AIM2 inflammasome activation and regulation: A structural perspective. J Struct Biol. 2017; 200(3): 279-82 [PMC5733693]
Selected Pre-FSU Publications
Yin Q, Fu TM, Li J, and Wu H. Structural Biology of Innate Immunity. Annu Rev Immunol. 2015; 33: 393-416 [DOI] [PMC4394028]
Li L, Yin Q, Kuss P, Maliga Z, Millán JL, Wu H, and Mitchison TJ. Hydrolysis of 2’3’-cGAMP by ENPP1 and design of non-hydrolyzable analogs. Nat Chem Biol. 2014; 10(12): 1043-8 [DOI] [PMC4232468]
Lu A, Magupalli VG, Ruan J, Yin Q, Atianand MK, Vos M, Schröder GF, Fitzgerald KA, Wu H and Egelman EH. Unified Polymerization Mechanism for the Assembly of ASC-Dependent Inflammasomes. Cell. 2014; 156(6): 1193-206 [DOI] [PMC4000066]
Yin Q, Sester DP, Tian Y, Hsiao YS, Lu A, Cridland JA, Sagulenko V, Thygesen SJ, Choubey D, Hornung V, Walz T, Stacey KJ and Wu H. Molecular Basis for p202-Mediated DNA Recognition and Interference with AIM2 Inflammasome Activation. Cell Reports. 2013; 4(2): 327-39 [DOI] [PMC3760141]
Yin Q#, Tian Y, Kabaleeswaran V, Jiang X, Tu D, Eck MJ, Chen ZJ, and Wu H. Cyclic di-GMP Sensing via the Innate Immune Signaling Protein STING. Mol Cell. 2012; 46(6): 735-45 (# Co-corresponding author) [DOI] [PMC3697849]
Yin Q, Lamothe B, Darnay BG and Wu H. Structural Basis for the Lack of E2 Interaction in the RING Domain of TRAF2. Biochemistry. 2009; 48(44): 10558-67 [DOI] [PMC2830148]
Yin Q, Lin SC, Lamothe B, Lu M, Lo YC, Hura G, Zheng Z, Rich RL, Campos AD, Myszka DG, Lenardo MJ, Darnay BG and Wu H. E2 interaction and dimerization in the crystal structure of TRAF6. Nat Struct Mol Biol. 2009; 16(6): 658-66 [DOI] [PMC2834951]
Yin Q, Park HH, Chang JY, Lin SC, Lo YC, da Graca LS, Jiang X, and Wu H. Caspase-9 Holoenzyme Is a Specific and Optimal Procaspase-3 Processing Machine. Mol Cell. 2006; 22(2): 259-68 [DOI] [PMC2904439]
Highlighted in Nat Rev Mol Cell Biol. 2006; 7(6): 389
Recommended by Faculty of 1000 http://f1000.com/1008194
Book Chapter
Wang B, Tian Y, and Yin Q. AIM2 Inflammasome Assembly and Signaling. Structural Immunology, edited by Jin T and Yin Q. Chapter 7, pp143-155
Yin Q, Lin SC, Lo YC, Damo SM and Wu H. Tumor Necrosis Factor Receptor- Associated Factors in Immune Receptor Signal Transduction. The Handbook of Cell Signaling, 2nd Ed, edited by R. A. Bradshaw and E. A. Dennis. 2010; Chapter 49, pp339-45
